MicroRNAs (miRNAs) are small non-coding RNAs that can regulate gene expression and that are frequently altered in human cancers. Computational approaches can help us understand how miRNAs are dysregulated and potentially offer new therapies. The dysregulated miRNAs can affect the hallmarks of cancer, such as sustaining proliferative signaling, evading growth suppressors, resisting cell death, activating invasion and metastasis, and inducing angiogenesis.
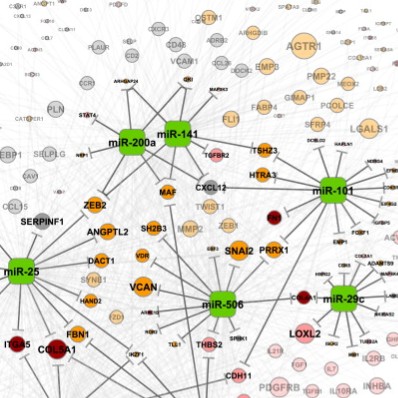
During the phase of The Cancer Genome Atlas (TCGA), our work on miRNAs focused on dysregulation of miRNA biogenesis and on a miRNA regulatory network associated with high-grade ovarian cancer. Currently, we are part of the NCI Genome Data Analysis Network (GDAN).
Our Center, operated jointly by ISB and British Columbia Cancer Agency (Dr. Steven Jones), is focused on integrated miRNA analysis of large datasets generated from NCI clinical trials. We are studying isoforms of miRNAs (IsomiRs, which exhibit length variation of miRNAs) that are associated with hallmark pathways of cancers. We are also interested in identifying circulating miRNAs as candidate biomarkers through integrative analysis of large-scale datasets, such as The Cancer Genome Atlas (TCGA) and External RNA Controls Consortium (ERCC). This approach may offer new diagnostic and prognostic options for cancer patients due to the stability of miRNAs in circulation and non-invasive measurement.
Publications
L. M. Moore, V. Kivinen, Y. Liu, M. Annala, D. Cogdell, X. Liu, C.G. Liu, R. Sawaya, O. Yli-Harja, I. Shmulevich, G.N. Fuller, W. Zhang, M. Nykter, “Transcriptome and small RNA deep sequencing reveals deregulation of miRNA biogenesis in human glioma,” The Journal of Pathology, Vol. 229, No. 3, pp. 449-459, 2013.
D. Yang, Y. Sun, L. Hu, H. Zheng, P. Ji, C. V. Pecot, Y. Zhao, S. Reynolds, H. Cheng, R. Rupaimoole, D. Cogdell, M. Nykter, R. Broaddus, C. Rodriguez-Aguayo, G. Lopez-Berestein, J. Liu, I. Shmulevich, A. K. Sood, K. Chen, W. Zhang, “Integrated Analyses Identify a Master MicroRNA Regulatory Network for the Mesenchymal Subtype in Serous Ovarian Cancer,” Cancer Cell, Vol. 23, No. 2, pp. 186-199, 2013.
G. Liu, Y. Sun, P. Ji, X. Li, D. Cogdell, D. Yang, B. C. Parker-Kerrigan, I. Shmulevich, K. Chen, A. K. Sood, F. Xue, W. Zhang, “MiR-506 Suppresses Proliferation and Induces Senescence by Directly Targeting the CDK4/6-FOXM1 Axis in Ovarian Cancer,” The Journal of Pathology, Vol. 233, No. 3, pp. 308-318, 2014.
Y. Sun, F. Guo, M. Bagnoli, F.-X. Xue, B.-C. Sun, I. Shmulevich, D. Mezzanzanica, K.-X. Chen, A. Sood, D. Yang, W. Zhang, “Key nodes of a microRNA network associated with the integrated mesenchymal subtype of high-grade serous ovarian cancer,” Chinese Journal of Cancer, Vol. 34, No. 1, pp. 28-40, 2015.
G. Liu, D. Yang, R. Rupaimoole, C. V. Pecot, Y. Sun, L. S. Mangala, X. Li, P. Ji, D. Cogdell, L. Hu, Y. Wang, C. Rodriguez-Aguayo, G. Lopez-Berestein, I. Shmulevich, L. De Cecco, K. Chen, D. Mezzanzanica, F. Xue, A. K. Sood, W. Zhang, “Augmentation of Response to Chemotherapy by microRNA-506 Through Regulation of RAD51 in Serous Ovarian Cancers,” Journal of the National Cancer Institute, Vol. 107, No. 7, djv108, 2015.


 shmulevich.isbscience.org/research/the-study-of-micrornas-in-cancer/
shmulevich.isbscience.org/research/the-study-of-micrornas-in-cancer/