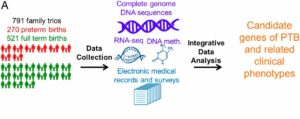
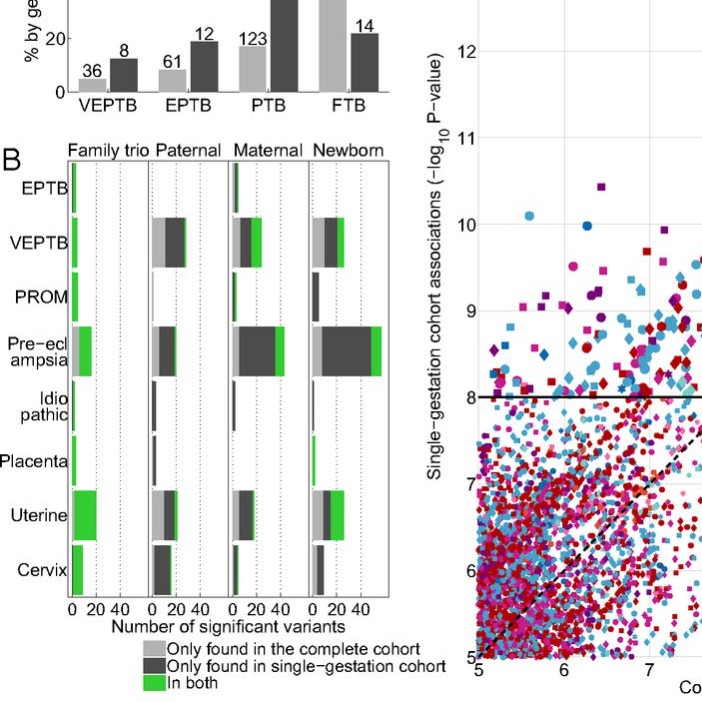
We have worked with Dr. John Niederhuber and his team at the Inova Translational Medicine Institute at Inova Health System on a large project on preterm birth (PTB). This work was with the collaboration of several scientists, such as Dr. Gustavo Glusman. The unique aspect of this study was the family-based design that included the parents and the newborn in the analysis. In this study, a family-based cohort of 791 family trios, including 270 PTB and 521 control families, was investigated by using whole-genome sequencing, RNA sequencing, and DNA methylation data [1].
The analysis of whole-genome sequencing data presented unique challenges and required the development of several methods and tools. We developed a method to use incomplete families in Family Based Association Test (FBAT) and evaluated its robustness against missing genotypes [2]. Using the large set of whole-genome sequencing data, we developed a method to accurately identify copy number variants [3]. The data for each parent and newborn also included demographic, clinical, and self-reported information, in addition to genomic, transcriptomic, and epigenetic data.
Statistically learning relationships from such heterogeneous data, when combined with genomic, transcriptomic, and epigenetic data, required methods that can scale to large feature sizes, capture the multivariate nature of statistical relationships, and be somewhat interpretable in the sense that it can be understood which features or combinations of features are most informative. Toward those ends, we developed a Random Forest implementation in GO that is highly scalable and efficient [4].
Publications
- T. A. Knijnenburg, J. G. Vockley, N. Chambwe, D. L. Gibbs, C. Humphries, K. C. Huddleston, E. Klein, P. Kothiyal, R. Tasseff, V. Dhankani, D. L. Bodian, W. S. W. Wong, G. Glusman, D. E. Mauldin, M. Miller, J. Slagel, S. Elasady, J. C. Roach, R. Kramer, K. Leinonen, J. Linthorst, R. Baveja, R. Baker, B. D. Solomon, G. Eley, R. K. Iyer, G. L. Maxwell, B. Bernard, I. Shmulevich, L. Hood, J. E. Niederhuber, “Genomic and molecular characterization of preterm birth,” Proceedings of the National Academy of Sciences of the USA, 201716314, 2019.
- V. Dhankani, D. L. Gibbs, T. Knijnenburg, R. Kramer, J. Vockley, J. Niederhuber, I. Shmulevich, B. Bernard, “Using incomplete trios to boost confidence in family based association studies,” Frontiers in Genetics, Vol. 7, No. 34, 2016.
- G. Glusman, A. Severson, V. Dhankani, M. Robinson, T. Farrah, D. Mauldin, A. B. Stittrich, S. A. Ament, J. Roach, M. Brunkow, D. Bodian, J. Vockley, I. Shmulevich, J. Niederhuber, L. Hood, “Identification of copy number variants in whole-genome data using Reference Coverage Profiles,” Frontiers in Genetics, Vol 6, No. 45, 2015.
- R. Bressler, R. B. Kreisberg, B. Bernard, J. E. Niederhuber, J. G. Vockley, I. Shmulevich, T. A. Knijnenburg, “CloudForest: A Scalable and Efficient Random Forest Implementation for Biological Data,” PLoS ONE, Vol. 10, No. 12, e0144820, 2015.


 shmulevich.isbscience.org/research/preterm-birth/
shmulevich.isbscience.org/research/preterm-birth/