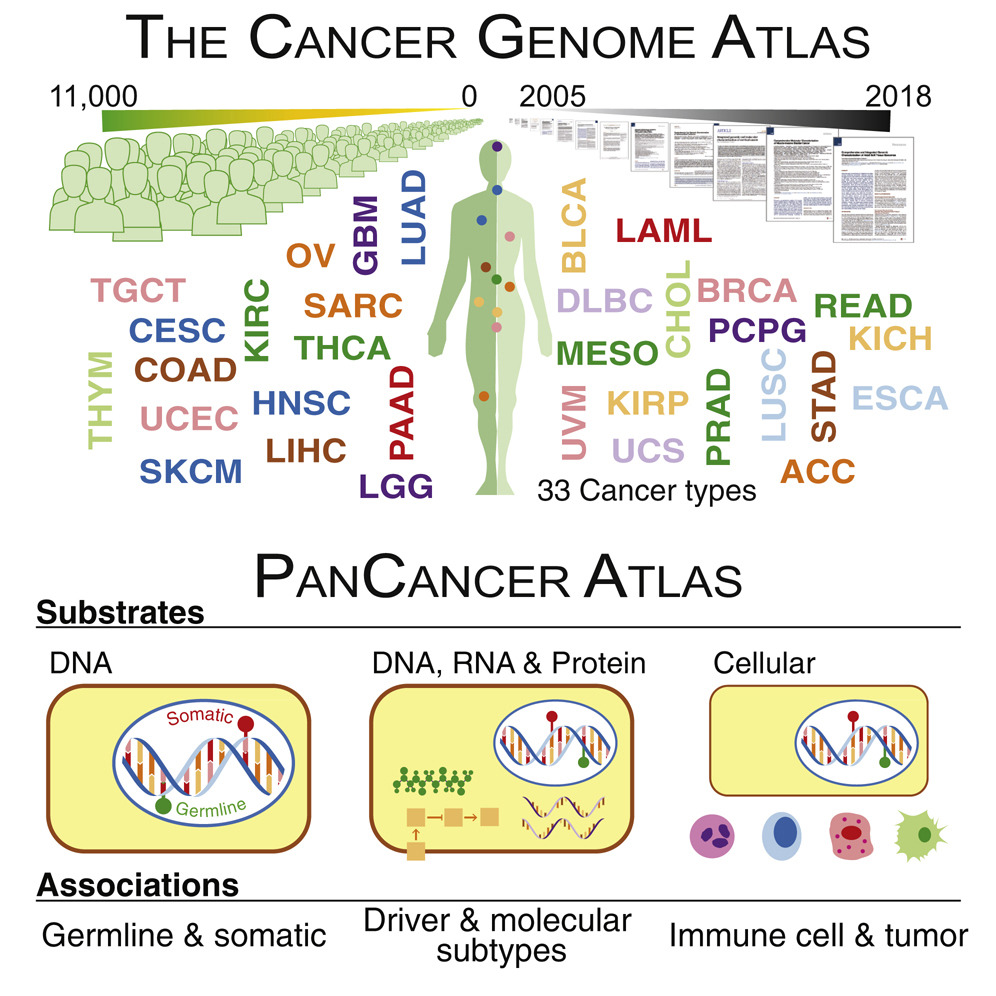
We describe our lab’s contributions to the Pan-Cancer Atlas consortium, which formed after the completion of The Cancer Genome Atlas (TCGA). The consortium generated 27 papers divided into three main categories: cell-of-origin patterns, oncogenic processes, and signaling pathways. Below, we describe our contributions.
In the area of Oncogenic Processes, we contributed to the flagship paper led by our collaborator Li Ding at Washington University which discussed the impact of genome alterations on the signaling and multi-omic profiles of human cancers as well as their influence on the tumor microenvironment.
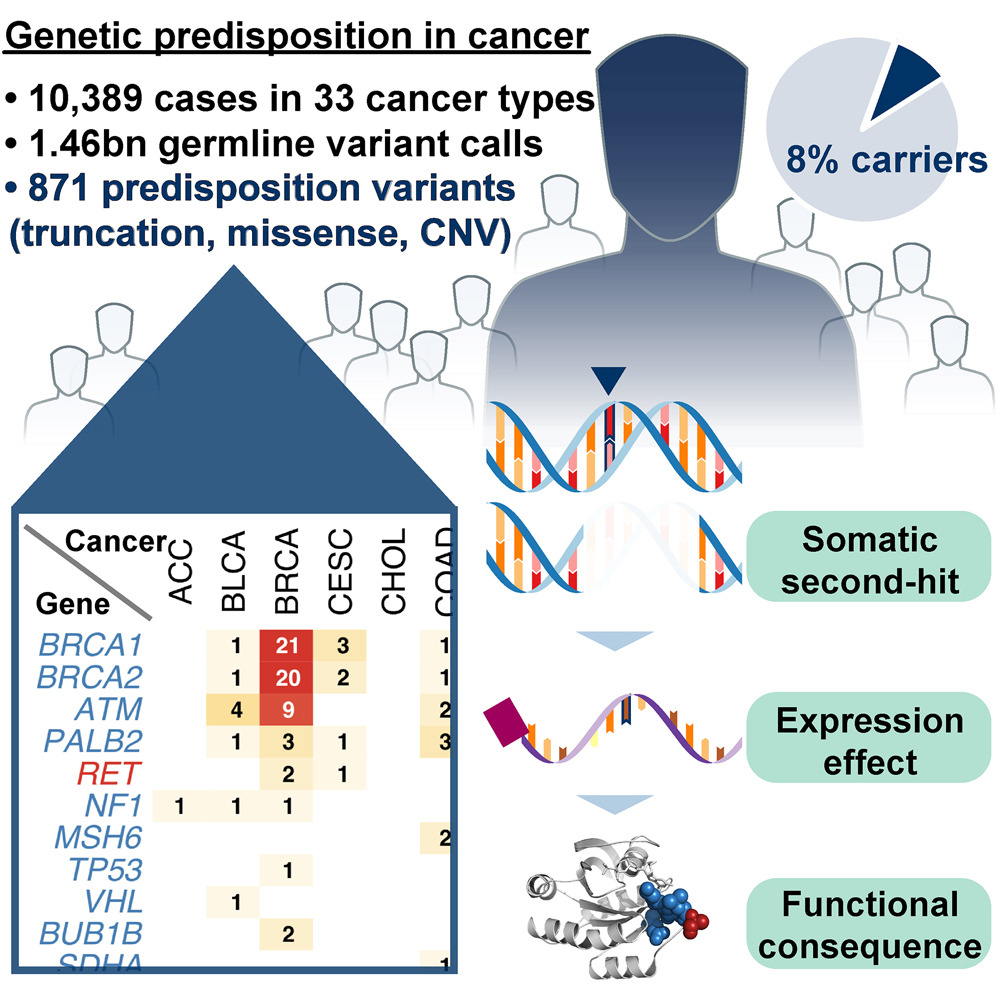
One unique aspect of TCGA data is the matched tumor and germline DNA sequence. This made it possible to analyze potentially pathogenic germline variants across all tumor types. This work and that of and several other Pan-Cancer Atlas working groups (fusion, splicing groups, and immune – below) involved massive computation that was made possible by the ISB Cancer Genomics Cloud (isb-cgc.org).
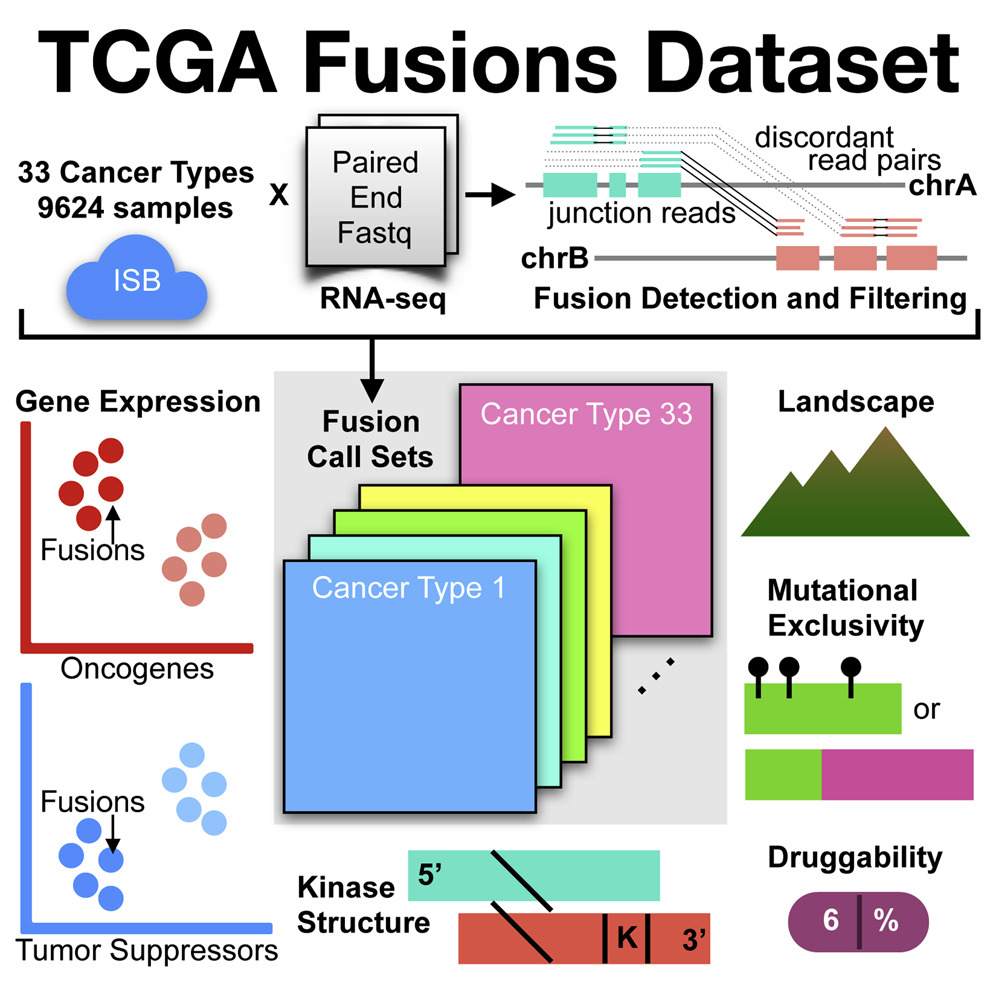
Another collaboration with the Ding lab and Dr. Matti Nykter (Tampere University), who is a former member of our lab, involved the analysis of gene fusion events and their patterns across tumor types.
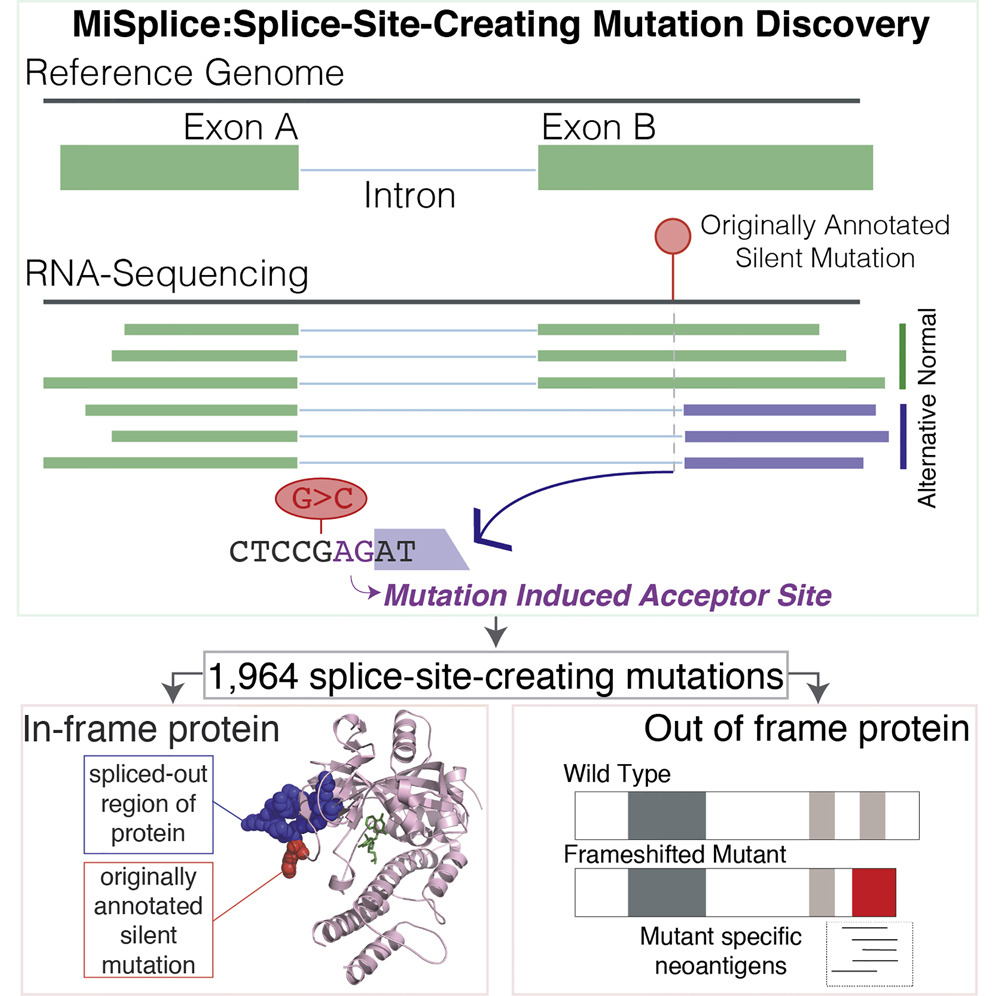
Our collaboration with the Ding Lab also included a study that identified nearly 2,000 splice-site-creating mutations (SCMs) from over 8,000 tumor samples across all 33 cancer types. Some of these mutations had been previously mis-annotated as missense and silent mutations. The study revealed that neoantigens induced by SCMs were significantly more immunogenic compared to missense mutations, underscoring the importance of characterizing the intratumoral immune response.
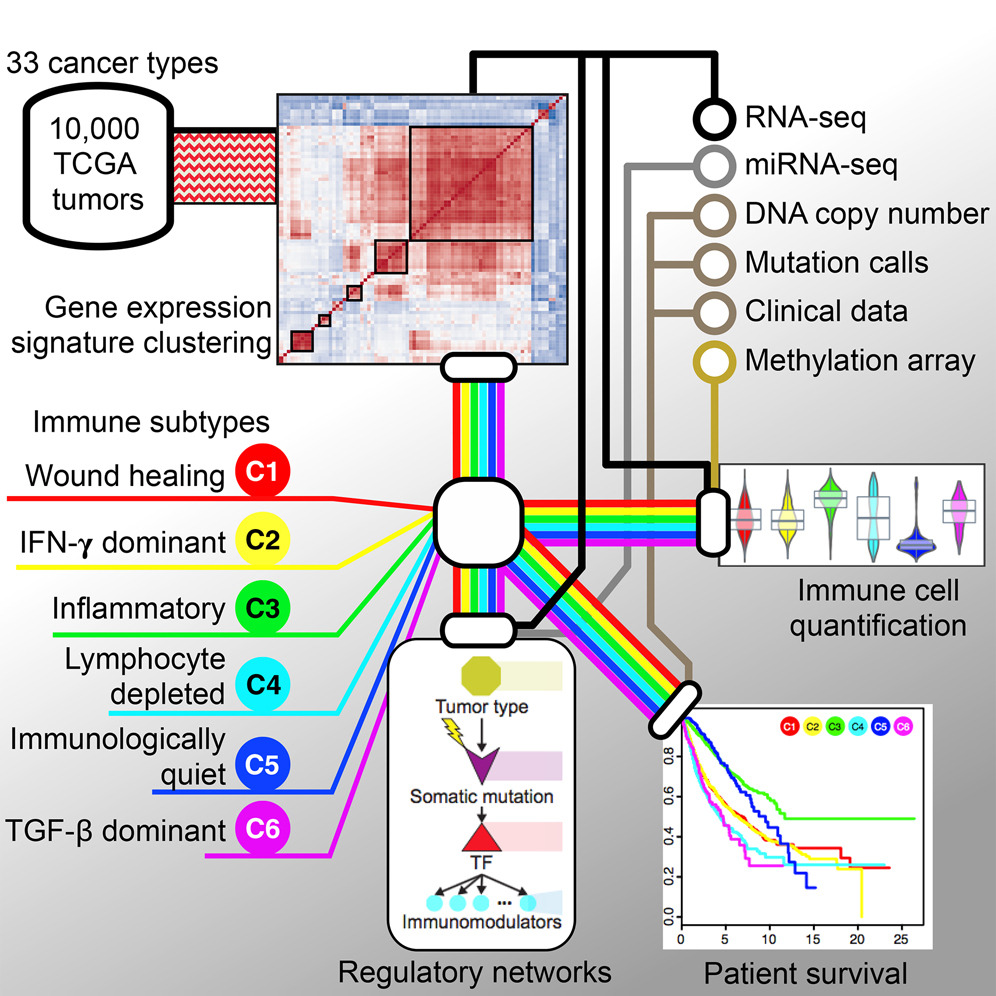
Our lab played a leading role in the Immune Response Working Group, co-chaired by Drs. Shmulevich, Thorsson, and close collaborator Dr. Benjamin Vincent at UNC, with key contributions from our lab member Dr. David Gibbs. This group performed a multifaceted immunogenomics analysis of more than 10,000 tumors, identifying six immune subtypes that encompass multiple cancer types and may define immune response patterns that are associated with prognosis. These and other analyses are available through the interactive CRI-iAtlas portal.
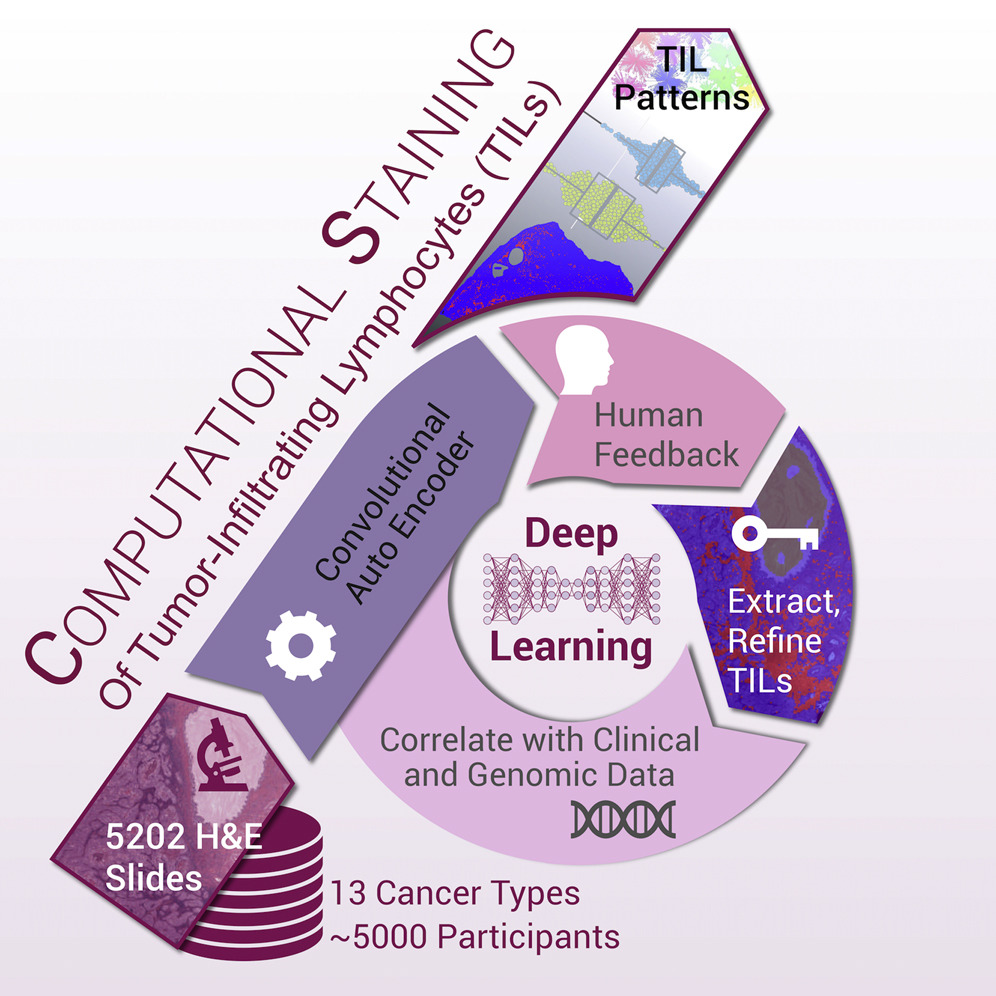
Related to the immune response study was another study, co-led by our lab member Dr. Vesteinn Thorsson, that identified tumor-infiltrating lymphocytes (TILs) from standard pathology cancer images (which is one of the data types available for each patient in TCGA, available through the ISB Cancer Genomics Cloud). Using a deep-learning method, this working group processed 5,202 digital images from 13 cancer types and correlated the resulting TIL spatial characteristics with TCGA data, relating TIL content to survival, tumor subtypes, and immune profiles.
In order to relate the molecular data from cancer patients to their clinical outcomes, it was important to harmonize all the clinical data, such as overall survival (OS) and progression-free interval (PFI) across 33 cancer types. We contributed to the analysis of clinicopathologic annotations of all cancer patients in the TCGA program and created a TCGA Clinical Data Resource.
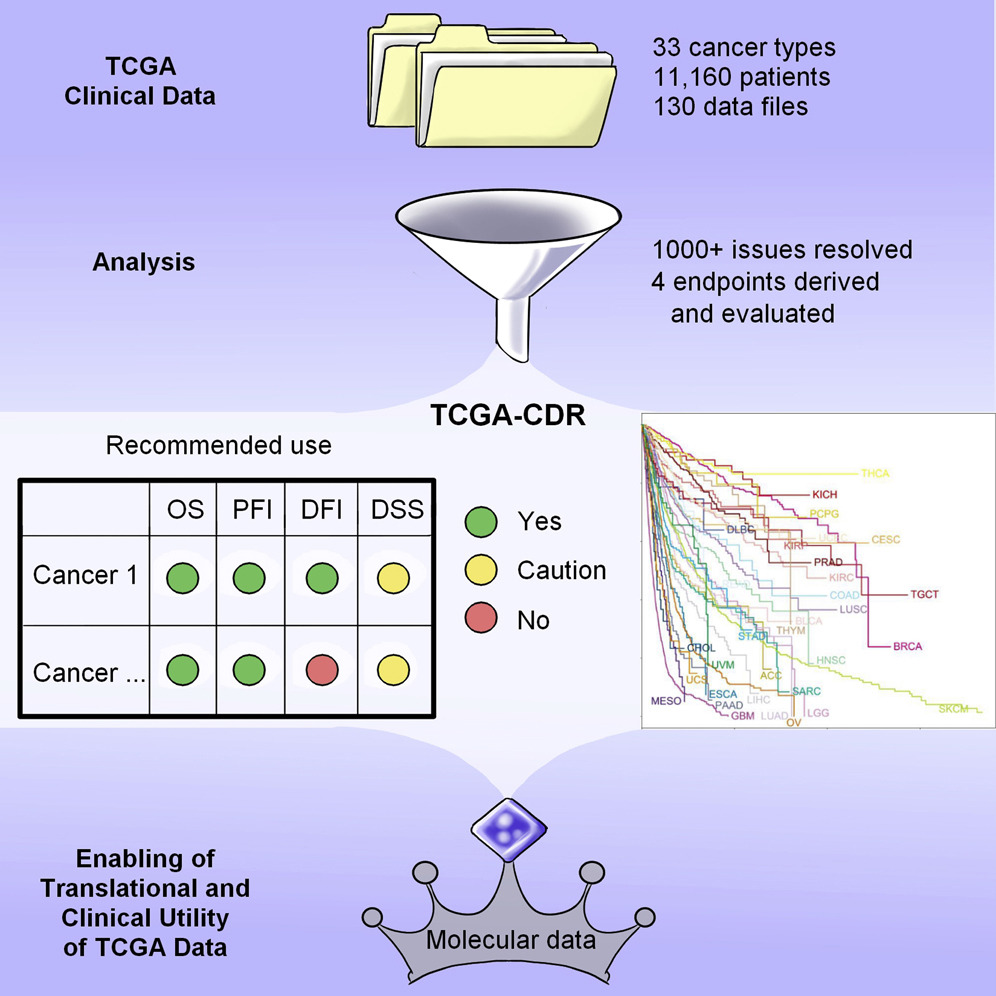
An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics
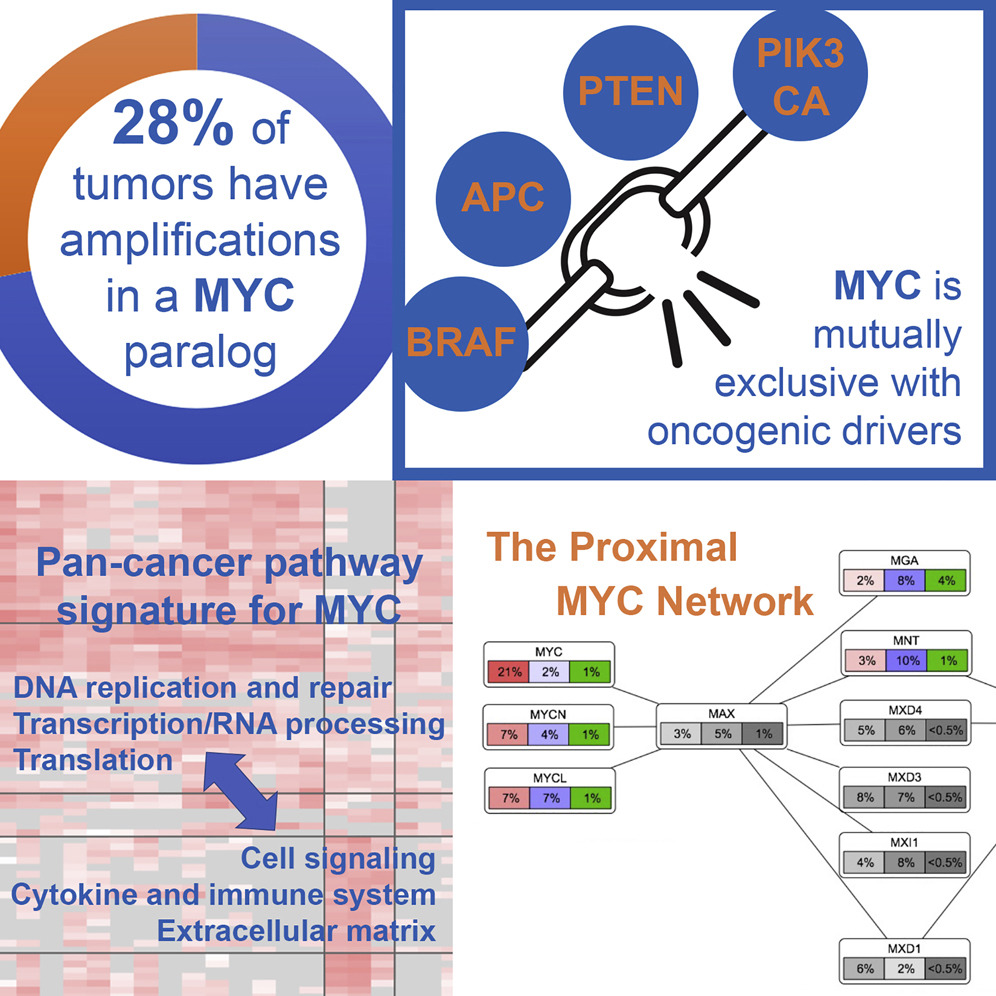
In the area of Signaling Pathways, we contributed to two studies. One study, co-led by our former lab member Dr. Brady Bernard (Earle A. Chiles Research Institute), performed an analysis of the frequency and extent of alterations of the MYC oncogene network across all 33 cancer types in TCGA.
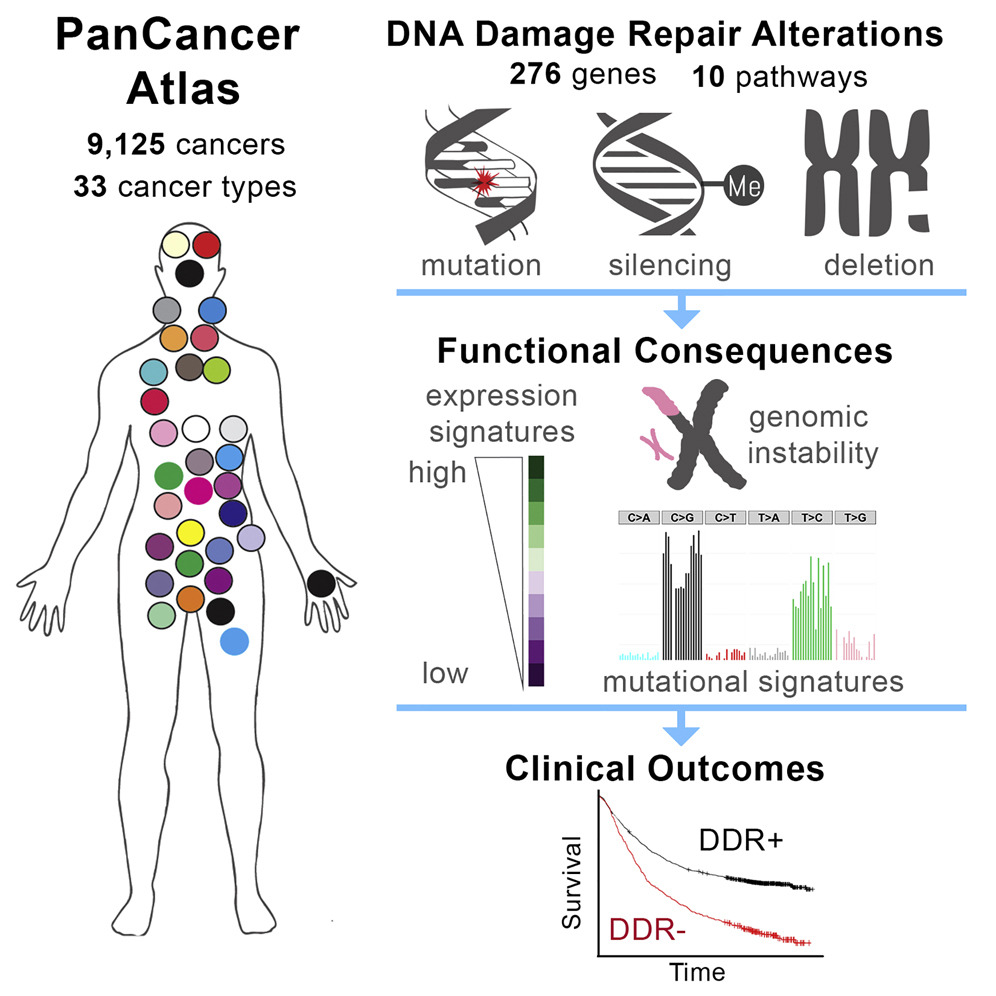
Another study, which was co-led by our close collaborator Dr. Raymond Monnat at the University of Washington (Genome Sciences and Pathology) and had two former lab members, Dr. Theo Knijnenburg (Allen Institute for Cell Science) and Dr. Nyasha Chambwe (St. Judes), performed a TCGA Pan-Cancer analysis of DNA damage repair (DDR) deficiency in cancer. This study used integrative analyses to identify frequent DDR alterations across 33 cancer types. These mutations may determine cancer progression and guide therapy.


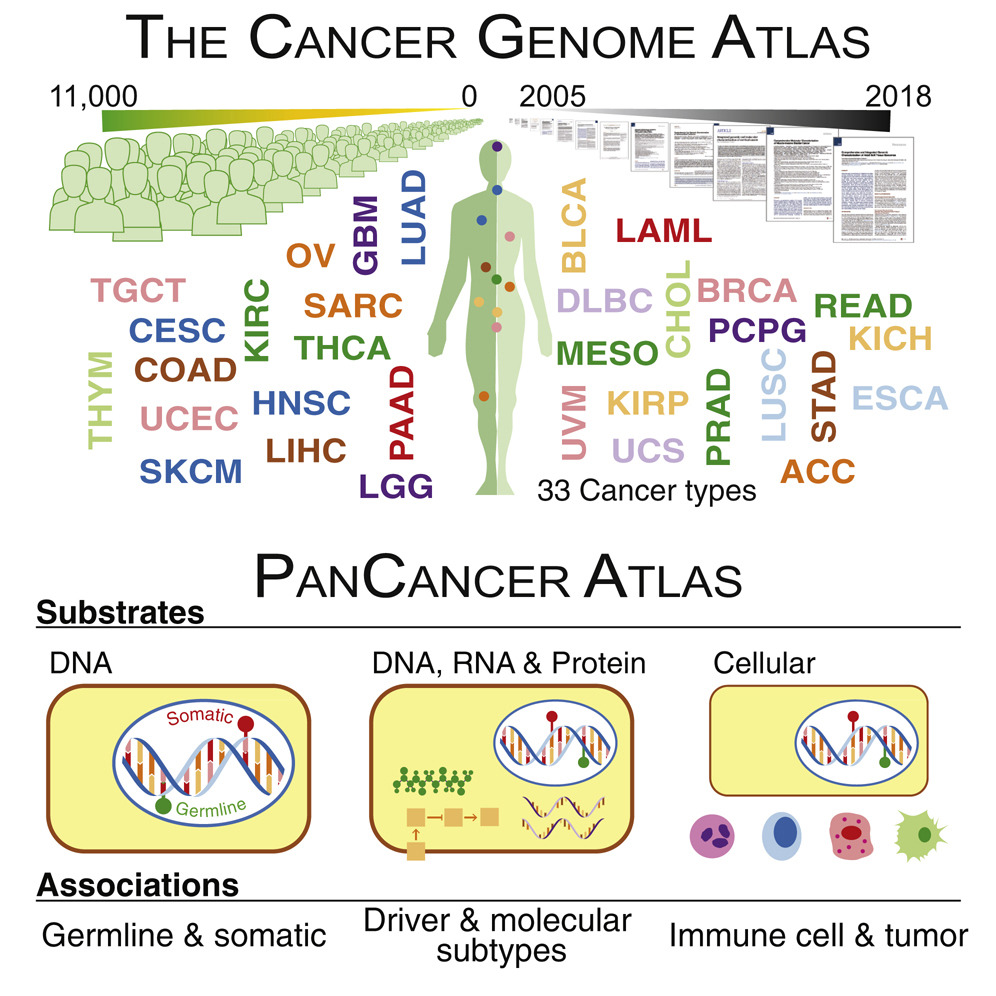
 shmulevich.isbscience.org/research/pan-cancer-atlas/
shmulevich.isbscience.org/research/pan-cancer-atlas/